

#PYMOL TUTORIAL MEASURE BOND DISTANCE FREE#
If what you want is some small molecule, you can try searching for the structure online, or can draw it using Avogadro, a free and open source molecule editor you can get at Visualizing and Exporting Models If you can't find your molecule, if it's not on the PDB it's probably because nobody knows it's structure yet. If you’re looking for cool structures and some explanations, the molecule of the month PDB articles at are a good way to start browsing. The biggest such database is the Protein Data Bank at
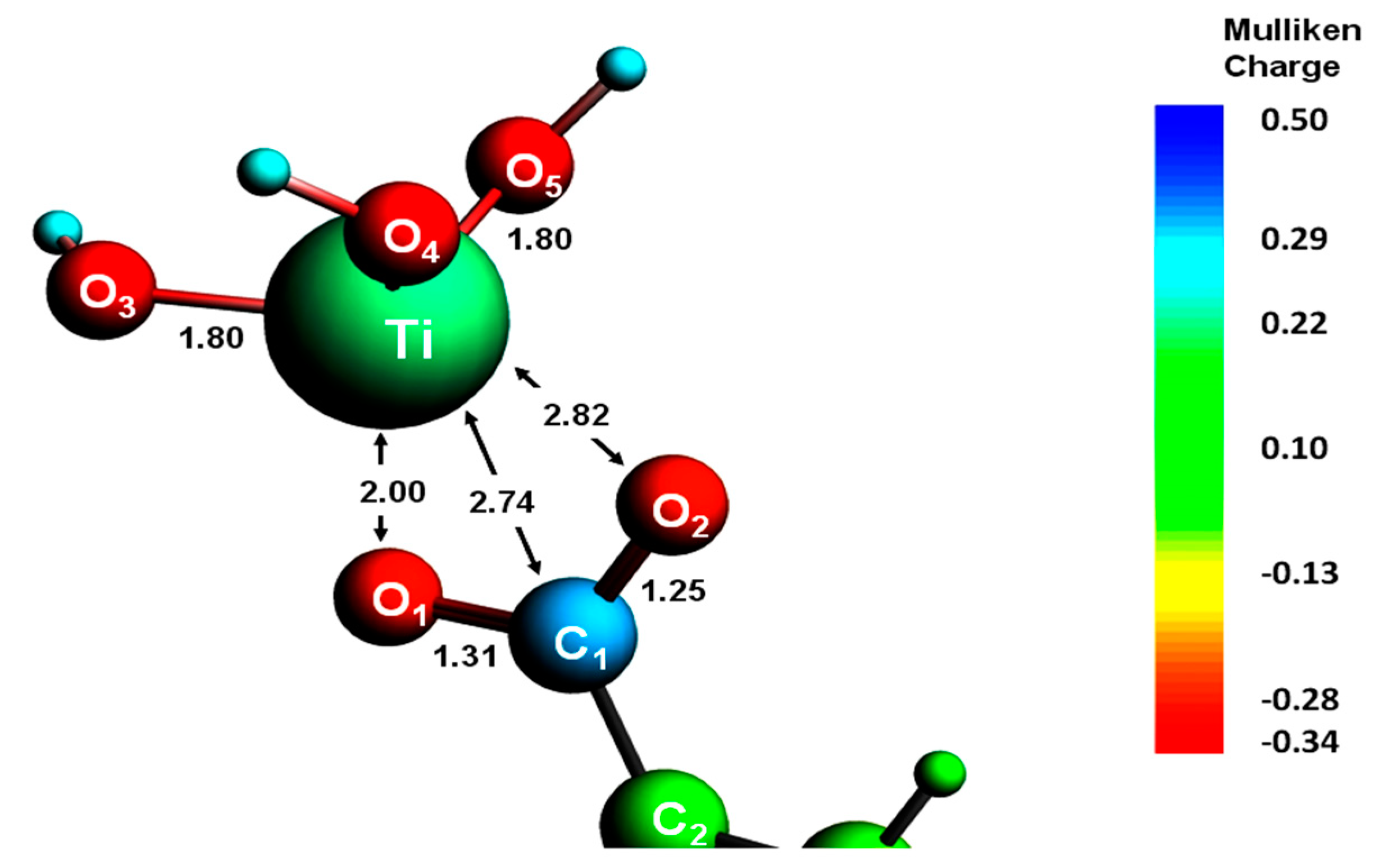
These structures are determined using an array of techniques such as x-ray crystallography and nuclear magnetic resonance, and when they are published they are also deposited in databases and become freely available for download.
The files containing the atoms coordinates for a molecule are called.


 0 kommentar(er)
0 kommentar(er)
